Deep convolutional neural network with 2% higher prediction accuracy for a 50-year-old problem of protein secondary structure prediction; in the last 20 years, 140 teams improved accuracy only by 3%; our improvement translates to 13 years of research with only 2 years of development; github.com/sh-maxim/ss
Assignment turn tool and library compiled with hierarchical DBSCAN and kMedoids clustering of 13 thousand of 3D protein fragments; 11 new types are discovered and 1 is eliminated; 2,700 downloads; github.com/sh-maxim/BetaTurn18
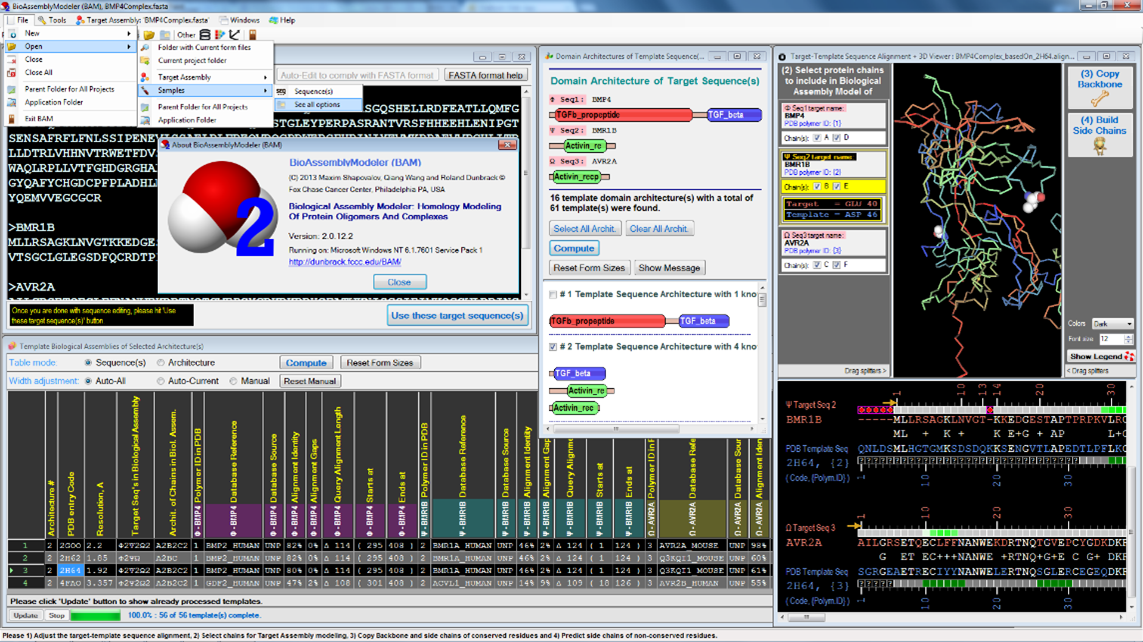
GUI application for 3D modeling of protein complexes; 2,200 users and 500 users of the first and second versions; dunbrack.fccc.edu/BAM
Cross-platform (Unix/Windows/Mac) program for atomic modeling of protein side chains; the most popular in the field; many commercial licenses; 9,000 users; dunbrack.fccc.edu/scwrl4
Statistically derived representation of protein side-chain atomic conformations, compiled from 4,000 protein structures using adaptive kernel density estimation and kernel regression of response variables; 900 users; dunbrack.fccc.edu/bbdep2010
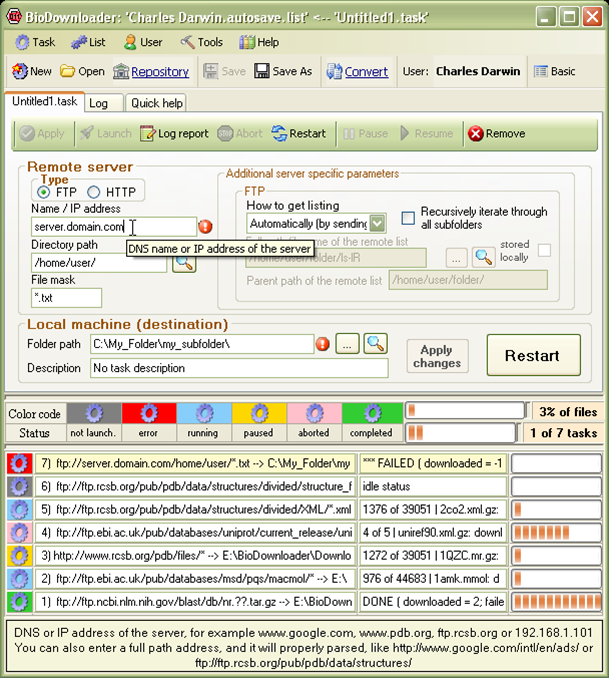
GUI application for downloading, updating and routine processing of biological files from FTP and HTTP servers; 650 users; dunbrack.fccc.edu/BioDownloader
Microsoft web server frontend for a Firebird database of protein interfaces. It supports different modes of querying and GUI browsing. For custom queries it allows submitting Linux jobs to a designated Linux server for additional computing and then retrieves results back.
Frontend of an SQLite database of antibody 3D conformations compiled with clustering.
Python Flask web application of kinase conformation database.
RHEL8, Apache web server for our research group content and product licensing.
[1] Adolf-Bryfogle, J., Labonte J., Kraft J., Shapovalov M., et al. Growing Glycans in Rosetta: Accurate de novo glycan modeling, density fitting, and rational sequon design. bioRxiv 2021.09.27.462000
[2] Shapovalov, M, Dunbrack, RL, Jr., and Vucetic, S. Multifaceted analysis of training and testing convolutional neural networks for protein secondary structure prediction. PLOS ONE, 2020. 15(5): e0232528.
[3] Shapovalov, M. Machine Learning Algorithms for Characterization and Prediction of Protein Structural Properties. Doctoral Dissertation, Temple University, 2019. Publication #: 20.500.12613/2356.
[4] Shapovalov, M, Vucetic, S, and Dunbrack, RL, Jr. A new clustering and nomenclature for beta turns derived from high-resolution protein structures. PLoS Comput Biol, 2019. 15(3): e1006844.
[5] Alford, R, Leaver-Fay, A, Jeliazkov, J, O'Meara, M, DiMaio, F, Park, H, Shapovalov, M, et al. The Rosetta All-Atom Energy Function for Macromolecular Modeling and Design. Journal of chemical theory and computation, 2017. 13(6): p. 3031-3048.
[6] Shapovalov, MV, Wang, Q, Xu, Q, Andrake, M, and Dunbrack Jr, RL. Bioassemblymodeler (BAM): User-friendly homology modeling of protein homo-and heterooligomers. PLOS ONE, 2014. 9(6): e98309.
[7] Shapovalov, MV and Dunbrack, RL, Jr. A smoothed backbone-dependent rotamer library for proteins derived from adaptive kernel density estimates and regressions. Structure, 2011. 19(6): p. 844-58.
[8] Ting, D, Wang, G, Shapovalov, M, Mitra, R, Jordan, MI, and Dunbrack, RL, Jr. Neighbor-dependent Ramachandran probability distributions of amino acids developed from a hierarchical Dirichlet process model. PLoS Comput Biol, 2010. 6(4): e1000763.
[9] Krivov, GG, Shapovalov, MV, and Dunbrack, RL, Jr. Improved prediction of protein side-chain conformations with SCWRL4. Proteins, 2009. 77(4): p. 778-95.
[10] Xu, Q, Canutescu, AA, Wang, G, Shapovalov, M, Obradovic, Z, and Dunbrack, RL, Jr. Statistical analysis of interface similarity in crystals of homologous proteins. J Mol Biol, 2008. 381(2): p. 487-507.
[11] Shapovalov, MV and Dunbrack, RL, Jr. Statistical and conformational analysis of the electron density of protein side chains. Proteins: Structure, Function and Genetics, 2007. 66(2): p. 279-303.
[12] Shapovalov, MV, Canutescu, AA, and Dunbrack, RL, Jr. BioDownloader: Bioinformatics downloads and updates in a few clicks. Bioinformatics, 2007.
[13] Shapovalov, M and Dunbrack Jr, RL. Non-parametric statistical analysis of the Ramachandran map in Biomolecular Forms and Functions: A Celebration of 50 Years of the Ramachandran Map. 2013, World Scientific. p. 76-94.
[14] Lehmann, A, Wixted, JH, Shapovalov, MV, Roder, H, Dunbrack, RL, Jr., and Robinson, MK. Stability engineering of anti-EGFR scFv antibodies by rational design of a lambda-to-kappa swap of the VL framework using a structure-guided approach. MAbs, 2015. 7(6): p. 1058-71.
[15] Berkholz, DS, Driggers, CM, Shapovalov, MV, Dunbrack, RL, Jr., and Karplus, PA. Nonplanar peptide bonds in proteins are common and conserved but not biased toward active sites. Proc Natl Acad Sci U S A, 2012. 109(2): p. 449-53.
[16] Beglov, D, Hall, DR, Brenke, R, Shapovalov, MV, Dunbrack, RL, Jr., Kozakov, D, and Vajda, S. Minimal ensembles of side chain conformers for modeling protein-protein interactions. Proteins, 2011.
[17] Shandler, SJ, Shapovalov, MV, Dunbrack, RL, Jr., and DeGrado, WF. Development of a rotamer library for use in beta-peptide foldamer computational design. J Am Chem Soc, 2010. 132(21): p. 7312-20.
[18] Loll, PJ, Derhovanessian, A, Shapovalov, MV, Kaplan, J, Yang, L, and Axelsen, PH. Vancomycin forms ligand-mediated supramolecular complexes. J Mol Biol, 2009. 385(1): p. 200-11.
[19] Berkholz, DS, Shapovalov, MV, Dunbrack, RL, Jr., and Karplus, PA. Conformation dependence of backbone geometry in proteins. Structure, 2009. 17(10): p. 1316-25.
[20] Karplus, PA, Shapovalov, MV, Dunbrack, RL, Jr., and Berkholz, DS. A forward-looking suggestion for resolving the stereochemical restraints debate: ideal geometry functions. Acta Crystallogr D Biol Crystallogr, 2008. 64(Pt 3): p. 335-6.
[21] Antoniou, I, Ivanov, VV, Kryanev, AV, Matokhin, VV, and Shapovalov, MV. On the efficient resources distribution in economics based on entropy. Physica A: Statistical Mechanics and its Applications, 2004. 336(3): p. 549-562.