-
Notifications
You must be signed in to change notification settings - Fork 46
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
internal fragmentation #82
Comments
|
I just implemented a first approach to the internal fragments problem (currently in another branch). calculateFragments("PQRST", type=c("b", "bIy"))
# mz ion type pos z seq
# 1 303.1775 bIy[2-3] bIy 2 1 QR
# 2 262.1510 bIy[3-4] bIy 3 1 RS
# 3 390.2096 bIy[2-4] bIy 2 1 QRS
# 4 270.1799 bIy[2-3]_ bIy_ 2 1 QR
# 5 229.1533 bIy[3-4]_ bIy_ 3 1 RS
# 6 357.2119 bIy[2-4]_ bIy_ 2 1 QRS
# 7 286.1510 bIy[2-3]* bIy* 2 1 QR
# 8 373.1830 bIy[2-4]* bIy* 2 1 QRSBecause of my minimal chemical background I am unsure whether all these calculations are correct and reasonable. I use the following additions: add <- c(a=-(mass["C"]+mass["O"]), # + H - CO
b=0, # + H
c=mass["N"]+3*mass["H"], # + H + NH3
x=mass["C"]+2*mass["O"], # + CO + OH
y=2*mass["H"]+mass["O"], # + H2 + OH
z=-(mass["N"]+mass["H"])+mass["O"], # - NH2 + OH
### internal fragments
aIx=mass["O"], # (- CO + CO) + OH
bIy=2*mass["H"]+mass["O"], # + H2 + OH
cIz=mass["H"]+mass["O"]) # + NH3 - NH2 + OH
## an additional H+ is added laterIs neutral loss resonable for (neutral loss is discussed in #47) |
|
@pavel-shliaha @sgibb any news on this front? |
|
The code is ready and could be merged if it is chemical correct. I am waiting for @pavel-shliaha's review. |
|
Ok, thanks. |
|
Any news on this front? |
|
I will play around with this shortly for proteins, once I finish the work on top-down with normal fragments. In the next few months |
|
@pavel-shliaha As far as I understand the internal fragments there could be all kind of combinations, e.g. |
|
Sorry, to keep you waiting, but I needed to submit the work we have already done. Yes it could be any sort of combination and yes having aIx, bIy and cIz is a good start. I think that top-down dataset we have is perfect for the work, since I expect more internal fragments than with small peptides. Can I ask you to implement the internal fragments in topdownr? This would allow me to test internal fragmentation much easier and more systematic and we need this functionality for top down work anyway. The internal fragments should also support adducts, since many of the fragments require adducts for identification. Just a reminder of the fragment types we observe, so if the probabilities of fragmenting a fragment is the same as fragmenting the precursor, then indeed bIy should be preferable in CID: |
|
Currently the internal fragment feature lives in an extra branch (a special side project) of install_github("lgatto/MSnbase@issue82-internalFragments")
install_github("sgibb/topdownr")Next you could simply add library("topdownr")
## default: fragments a, b, c, x, y, z
tdsDflt <- readTopDownFiles(topdownrdata::topDownDataPath("myo"))
tdsDflt
# TopDownSet object (4.33 Mb)
# - - - Protein data - - -
# Amino acid sequence (153): GLSDGEWQQVLNVWGKVEADIAGHGQ...GAMTKALELFRNDIAAKYKELGFQG
# Mass : 16964.96
# Modifications (3): Carbamidomethyl, Acetyl, Met-loss
# - - - Fragment data - - -
# Number of theoretical fragments: 2700
# Theoretical fragment types (18): a, a_, a*, b, b_, ..., y_, y*, z, z_, z*
# Theoretical mass range: [68.03;16925.98]
# - - - Condition data - - -
# Number of conditions: 1852
# Number of scans: 5882
# Condition variables (61): File, Scan, ..., Sample, MedianIonInjectionTimeMs
# - - - Intensity data - - -
# Size of array: 2700x5882 (0.67% != 0)
# Number of matched fragments: 106282
# Intensity range: [109.29;10704001.00]
# - - - Processing information - - -
# [2018-06-03 20:41:53] 106282 fragments [2700;5882] matched (tolerance: 5 ppm, strategies ion/fragment: remove/remove).
# [2018-06-03 20:41:53] Condition names updated based on: Mz, AgcTarget, EtdReagentTarget, EtdActivation, CidActivation, HcdActivation. Order of conditions changed. 1852 conditions.
# [2018-06-03 20:41:54] Recalculate median injection time based on: Mz, AgcTarget.
## internal fragments
tdsIntF <- readTopDownFiles(topdownrdata::topDownDataPath("myo"),
type=c("a", "b", "c", "x", "y", "z",
"aIx", "bIy", "cIz"))
tdsIntF
# TopDownSet object (24.70 Mb)
# - - - Protein data - - -
# Amino acid sequence (153): GLSDGEWQQVLNVWGKVEADIAGHGQ...GAMTKALELFRNDIAAKYKELGFQG
# Mass : 16964.96
# Modifications (3): Carbamidomethyl, Acetyl, Met-loss
# - - - Fragment data - - -
# Number of theoretical fragments: 102999
# Theoretical fragment types (27): a, a_, a*, aIx, aIx_, ..., y_, y*, z, z_, z*
# Theoretical mass range: [68.03;16925.98]
# - - - Condition data - - -
# Number of conditions: 1852
# Number of scans: 5882
# Condition variables (61): File, Scan, ..., Sample, MedianIonInjectionTimeMs
# - - - Intensity data - - -
# Size of array: 102999x5882 (0.09% != 0)
# Number of matched fragments: 515232
# Intensity range: [71.40;10704001.00]
# - - - Processing information - - -
# [2018-06-03 20:42:20] 515232 fragments [102999;5882] matched (tolerance: 5 ppm, strategies ion/fragment: remove/remove).
# [2018-06-03 20:42:20] Condition names updated based on: Mz, AgcTarget, EtdReagentTarget, EtdActivation, CidActivation, HcdActivation. Order of conditions changed. 1852 conditions.
# [2018-06-03 20:42:20] Recalculate median injection time based on: Mz, AgcTarget.
## rowViews
rowViews(tdsDflt)
# FragmentViews on a 153-letter sequence:
# GLSDGEWQQVLNVWGKVEADIAGHGQEVLIRLFTGHPE...SKHPGDFGADAQGAMTKALELFRNDIAAKYKELGFQG
# Mass:
# 16964.964625
# Modifications:
# Carbamidomethyl
# Acetyl
# Met-loss
# Views:
# start end width mass name type z
# [1] 1 1 1 68.03 z1_ z_ 1 [G]
# [2] 1 1 1 72.04 a1 a 1 [G]
# [3] 1 1 1 85.05 y1_ y_ 1 [G]
# [4] 1 1 1 100.04 b1 b 1 [G]
# [5] 1 1 1 101.02 z1 z 1 [G]
# ... ... ... ... ... ... ... ... ...
# [2696] 2 153 152 16893.90 x152* x* 1 [LSDGEWQQVLNVWG...NDIAAKYKELGFQG]
# [2697] 1 152 152 16908.95 b152 b 1 [GLSDGEWQQVLNVW...RNDIAAKYKELGFQ]
# [2698] 1 152 152 16908.95 c152* c* 1 [GLSDGEWQQVLNVW...RNDIAAKYKELGFQ]
# [2699] 2 153 152 16910.93 x152 x 1 [LSDGEWQQVLNVWG...NDIAAKYKELGFQG]
# [2700] 1 152 152 16925.98 c152 c 1 [GLSDGEWQQVLNVW...RNDIAAKYKELGFQ]
rowViews(tdsIntF)
# FragmentViews on a 153-letter sequence:
# GLSDGEWQQVLNVWGKVEADIAGHGQEVLIRLFTGHPE...SKHPGDFGADAQGAMTKALELFRNDIAAKYKELGFQG
# Mass:
# 16964.964625
# Modifications:
# Carbamidomethyl
# Acetyl
# Met-loss
# Views:
# start end width mass name type z
# [1] 1 1 1 68.03 z1_ z_ 1 [G]
# [2] 1 1 1 72.04 a1 a 1 [G]
# [3] 1 1 1 85.05 y1_ y_ 1 [G]
# [4] 73 74 2 98.05 aIx[73-74]_ aIx_ 1 [GG]
# [5] 73 74 2 99.06 cIz[73-74]_ cIz_ 1 [GG]
# ... ... ... ... ... ... ... ... ...
# [102995] 2 153 152 16893.90 x152* x* 1 [LSDGEWQQVL...AKYKELGFQG]
# [102996] 1 152 152 16908.95 b152 b 1 [GLSDGEWQQV...AAKYKELGFQ]
# [102997] 1 152 152 16908.95 c152* c* 1 [GLSDGEWQQV...AAKYKELGFQ]
# [102998] 2 153 152 16910.93 x152 x 1 [LSDGEWQQVL...AKYKELGFQG]
# [102999] 1 152 152 16925.98 c152 c 1 [GLSDGEWQQV...AAKYKELGFQ]
## select only internal fragments
tdsIntF[c("aIx", "bIy", "cIz"),]
# TopDownSet object (9.74 Mb)
# - - - Protein data - - -
# Amino acid sequence (153): GLSDGEWQQVLNVWGKVEADIAGHGQ...GAMTKALELFRNDIAAKYKELGFQG
# Mass : 16964.96
# Modifications (3): Carbamidomethyl, Acetyl, Met-loss
# - - - Fragment data - - -
# Number of theoretical fragments: 33975
# Theoretical fragment types (3): aIx, bIy, cIz
# Theoretical mass range: [131.05;16827.93]
# - - - Condition data - - -
# Number of conditions: 1852
# Number of scans: 5882
# Condition variables (61): File, Scan, ..., Sample, MedianIonInjectionTimeMs
# - - - Intensity data - - -
# Size of array: 33975x5882 (0.07% != 0)
# Number of matched fragments: 148392
# Intensity range: [71.40;1966595.88]
# - - - Processing information - - -
# [2018-06-03 20:42:20] 515232 fragments [102999;5882] matched (tolerance: 5 ppm, strategies ion/fragment: remove/remove).
# [2018-06-03 20:42:20] Condition names updated based on: Mz, AgcTarget, EtdReagentTarget, EtdActivation, CidActivation, HcdActivation. Order of conditions changed. 1852 conditions.
# [2018-06-03 20:42:20] Recalculate median injection time based on: Mz, AgcTarget.
# [2018-06-03 20:47:51] Subsetted 515232 fragments [102999;5882] to 148392 fragments [33975;5882]. |
|
|
|
On @pavel-shliaha's request I revert the changes from To test this feature you have to install both packages from specific branches: devtools::install_github("lgatto/MSnbase@issue82-internalFragments")
devtools::install_github("sgibb/topdownr@internalFragments") |
|
What's the status of this? |
|
It is based on my limited theoretical knowledge about internal fragments and is
not tested/verified by @pavel-shliaha or anyone else.
Quoting Laurent Gatto (2020-10-28 22:48:08)
… What's the status of this?
--
You are receiving this because you were mentioned.
Reply to this email directly or view it on GitHub:
#82 (comment)
|
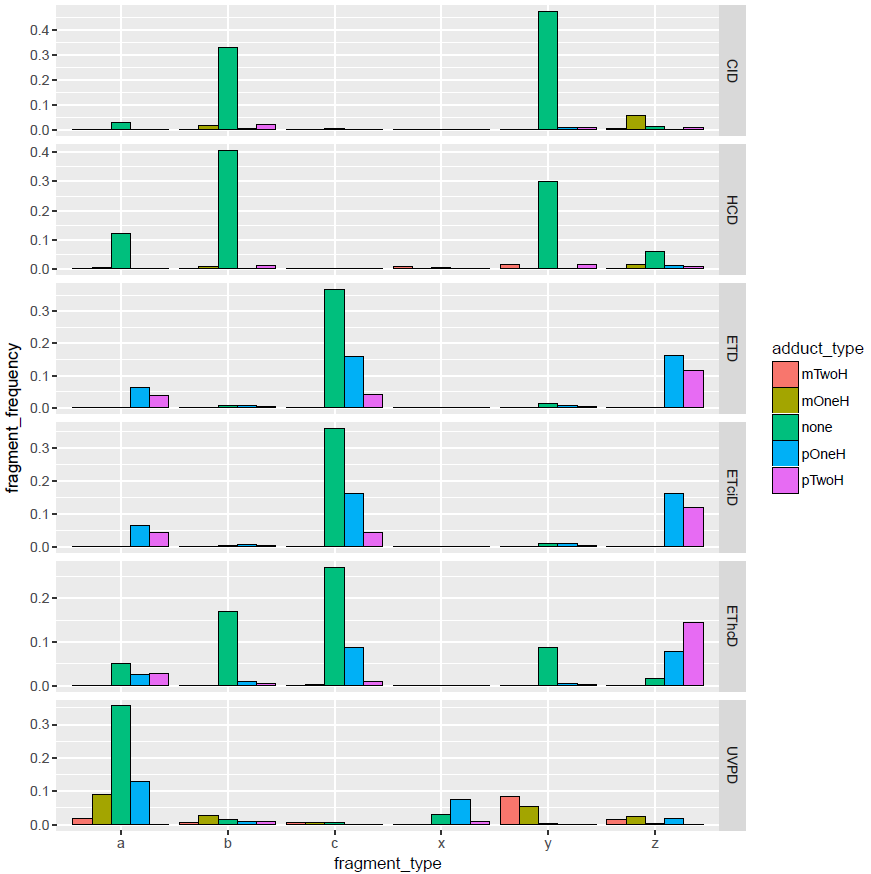
I am now doing some intact protein analysis and it was recently demonstrated that when you fragment proteins you produce a lot of internal fragments:
http://www.ncbi.nlm.nih.gov/pubmed/25716753
considering these internal fragments results in a huge boost in coverage. Could internal fragmentation be introduced in calculateFragments
The text was updated successfully, but these errors were encountered: