-
Notifications
You must be signed in to change notification settings - Fork 16
MRI_Neurite_Analyzer
The toolset helps to segment neurites, measure the distances on the neurites to the closest soma, assign each neurite to a soma and to measure the FISH-signal on the neurites.

You must have the ilastik4ij-plugin and the Neurite_Labelling_IJ_Plugin installed. Save the toolset neurite_analyzer.ijm into the macros/toolsets-folder of your FIJI installation. Download and unzip the ilastik classifier and the training images. Unzip the archive in the macros/toolbars-folder. Select neurite_analyzer from the >>-button:
Before using the toolset, please configure the ilastik4ij-plugin. Run Plugins>ilastik>configure ilastik executable location and set the path to your ilastik-executable.
- the first button opens this help page
-
n-button: segment the nuclei in the active image -
s-button: segment the neurites in the active image (this might take a while) -
d-button: calculate for each point on a neurite, the length of the shortest path along the neurites to the closest soma -
l-button: label each point with the label of the closest soma following a path on the neurites -
f-button: detect the FISH spots and measures the distance along a neurite to the closest soma and the label of the neurite -
bn-button: batch segment the nuclei, segement the nuclei in all the images in all direct subfolders of the input folder -
h5-button: batch export the neurite images as h5-images, optionally apply histogram equalization -
i-button: the button does nothing, but remind the user to use ilastik for the neurite segmentation -
bs-button: batch convert the output of the ilastik segmentation into ImageJ rois -
bd-button: batch calculate the distances and the neurite labelling -
bf-button: batch detect the FISH spots and measure the label and the distance to the soma -
b-button: run all the batch steps after having segmented the neurites with ilastik, i.e.bn,bs,bdandbf.
Nuclei are segmented by applying a Gaussian blur filter, followed by an auto-threshold. The particle analyzer is then used to create rois.
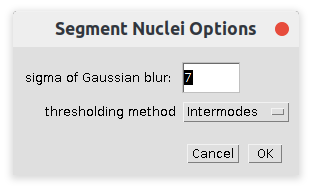
Right-click the n-button to open the options-dialog:
- sigma of Gaussian blur
- The sigma of the Gaussian blur filter. Select a sigma that just makes the inner structures of the nuclei disappear.
- Thresholding method
- The auto-thresholding method used to segment the nuclei
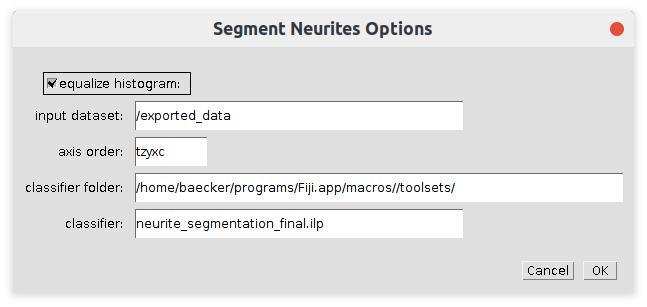
The neurites are segmented using a pretrained ilastik-classifier. The execution of this operation may be long. It might be preferable to use ilastik directly for the neurite segmentation. You can use the provided ilastik classifier or create your own.
- equalize histogram
- If selected a histogram equalization is run on the image before the classifier is applied. The equalization should be used consistently between training and application
- /exported_data
- The dataset within the h5-file to be used
- axis order
- The meaning of the different dimensions of the dataset.
- classifier folder
- The path to the folder containing the classifier (ilp-file)
- classifier
- The trained ilastik classifier. The training images must available at the relative paths stored in the classifier


 Volker Bäcker
Volker Bäcker